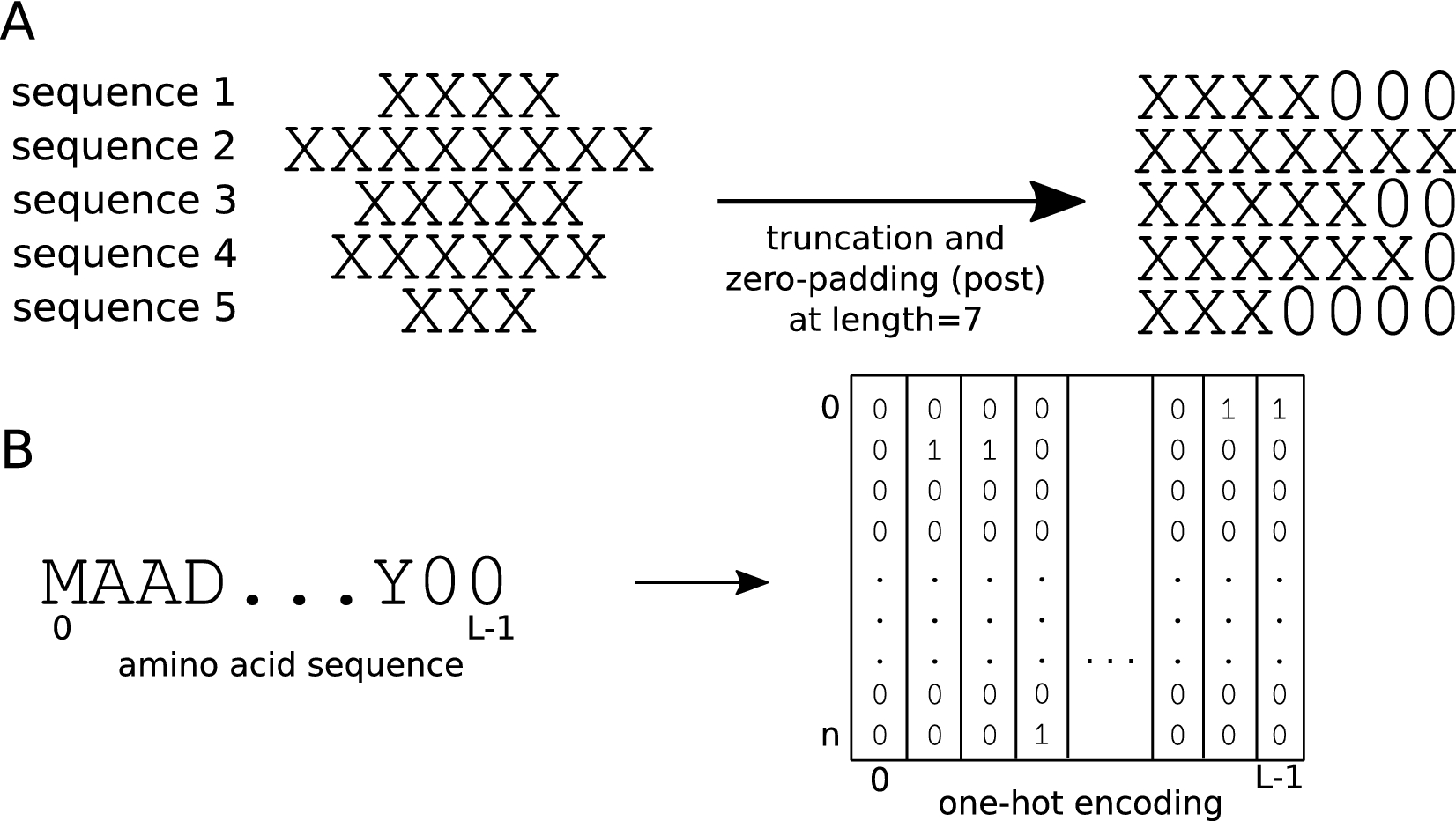
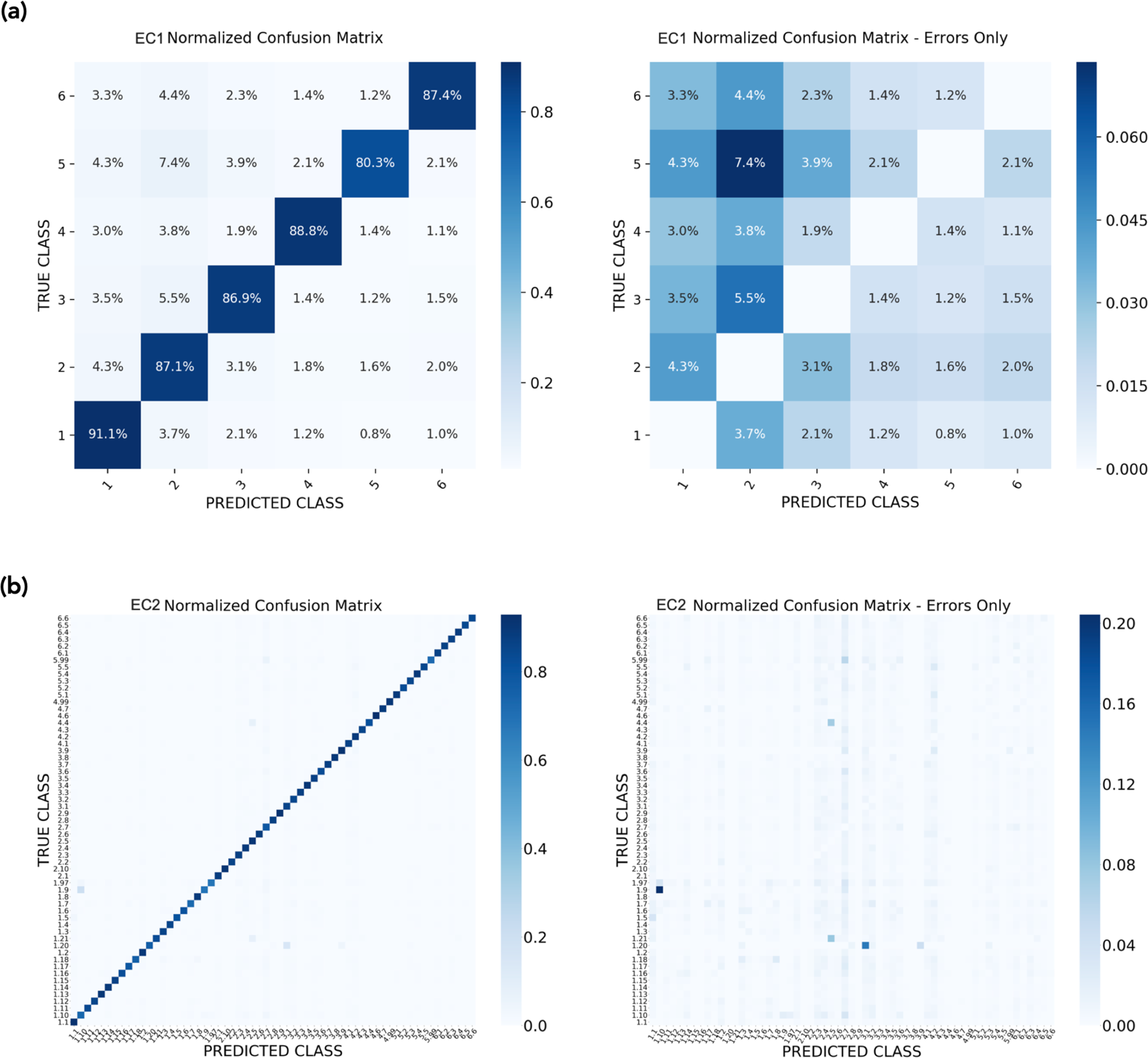
Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction
$ 18.99 · 4.9 (134) · In stock

MECE: a method for enhancing the catalytic efficiency of glycoside hydrolase based on deep neural networks and molecular evolution - ScienceDirect

Machine learning methods for predicting protein structure from single sequences - ScienceDirect

Unified rational protein engineering with sequence-only deep representation learning
Deep learning of a bacterial and archaeal universal language of life enables transfer learning and illuminates microbial dark matter

Deep learning program to predict protein functions based on sequence information - ScienceDirect

PDF) Zero-Padding and Spatial Augmentation-Based Gas Sensor Node Optimization Approach in Resource-Constrained 6G-IoT Paradigm

Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction

DeepRCI: predicting RNA-chromatin interactions via deep learning with multi-omics data

Deep neural language modeling enables functional protein generation across families

Amino Acid Encoding Methods for Protein Sequences: A Comprehensive Review and Assessment

Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction

IJMS, Free Full-Text